Bioanalytical Facility
The department hosts a custom built, well-equipped analytical facility for proteomic workflows for protein and peptide analysis and characterisation of nucleic acids, nucleosides and other metabolites.
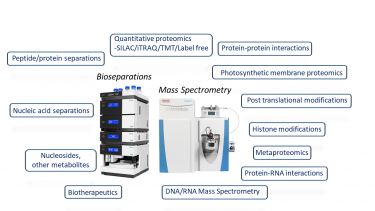
Overview
Our expertise is bioanalysis of multiple cell systems from mammalian to algae and bacteria; working on projects within the Biological Engineering research theme.
We also collaborate on range of life science projects across faculties (Health, Science) and with external academic and industry partners. Our collaborations include other University of Sheffield schools, including:
- School of Biosciences
- School of Medicine and Population Health
- School of Clinical Dentistry
- Sheffield Institute for Translational Neuroscience
Our external collaborators include Professor Edze Westra, University of Exeter, Professor Josef Komenda and Dr Roman Sobotka, The Czech Academy of Sciences
Industrial collaborators include Syngenta, Glaxo Smith Kline, ThermoFisher, FUJIFILM Diosynth Biotechnologies and AstraZeneca.
Research areas
- Quantitative proteomics - SILAC, metabolic labeling, iTRAQ and label free quantification
- Protein-protein, Protein-RNA interactions - quantitative analysis of protein-protein interactions and the mRNA interactome
- Protein post translational modifications - phosphorylation, methylation, glycosylation and histone PTMs
- Characterisation of biotherapeutic biomolecules - biopharmaceuticals, RNA therapeutics
- Nucleic Acid Mass Spectrometry
- Metaproteomics
Equipment
- Q Exactive™ HF hybrid quadrupole-Orbitrap mass spectrometer (Thermo Scientific)-BBSRC funded
- maXis ultra high ToF instrument (Bruker Daltonics))
- amaZon Ion trap (Bruker Daltonics)
- Thermo Finnigan TRACE DSQ GC-MS System
Bioseparations
- Agilent 7890A gas - chromatograph (GC) instrument equipped with a flame ionization detector
- A wide range of HPLC/UPLC systems covering analytical, capillary and nano flow rates (Agilent, Thermo Scientific)
Publications
- Quantitative proteomics
-
Kaneva IN, Longworth J, Sudbery PE & Dickman MJ (2018) Quantitative Proteomic Analysis in Candida albicans Using SILAC-Based Mass Spectrometry. Proteomics, 18(5-6), 1700278-1700278. View this article in WRRO
MacGregor-Chatwin C, Jackson PJ, Sener M, Chidgey JW, Hitchcock A, Qian P, Mayneord GE, Johnson MP, Luthey-Schulten Z, Dickman MJ, Scanlan DJ (2019). Membrane organization of photosystem I complexes in the most abundant phototroph on Earth. Nature Plants 5(8):879-89. View this article in WRRO
Krynicka V, Georg J, Jackson PJ, Dickman MJ, Hunter CN, Futschik ME, Hess WR, Komenda J. (2019). Depletion of the FtsH1/3 proteolytic complex suppresses the nutrient stress response in the cyanobacterium Synechocystis PCC6803. The Plant Cell doi.org/10.1105/tpc.19.00411.
Jackson PJ, Lewis HJ, Tucker JD, Hunter CN & Dickman MJ (2012) Quantitative proteomic analysis of intracytoplasmic membrane development in Rhodobacter sphaeroides.. Mol Microbiol, 84(6), 1062-1078.
Raut MP, Couto N, Pham TK, Evans C, Noirel J, Wright PC (2016) Quantitative proteomic analysis of the influence of lignin on biofuel production by Clostridium acetobutylicum ATCC 824. Biotechnol Biofuels 9: 113
Corfe BM, Majumdar D, Assadsangabi A, Marsh AMR, Cross SS, Connolly JB, Evans CA & Lobo AJ (2015) Inflammation decreases keratin level in ulcerative colitis; inadequate post-inflammatory restoration associates with increased risk of colitis-associated-cancer. BMJ Gastroenterology 2: e000024.
- Protein-protein, Protein-RNA interactions
-
Chidgey JW, Jackson PJ, Dickman MJ & Hunter CN (2017). PufQ regulates porphyrin flux at the haem/bacteriochlorophyll branchpoint of tetrapyrrole biosynthesis via interactions with ferrochelatase. Mol. Microbiol. 106: 961-975.
Chidgey JW, Linhartova M, Komenda J, Jackson PJ, Dickman MJ, Canniffe DP, Konik P, Pilny J, Hunter CN & Sobotka R (2014). A cyanobacterial chlorophyll synthase-HliD complex associates with the Ycf39 protein and the YidC/Alb3 insertase. The Plant Cell 26: 1267-1279.
Hollingshead S, Kopecna J, Jackson PJ, Canniffe DP, Davison PA, Dickman MJ, Sobotka R & Hunter CN (2012) Conserved chloroplast open-reading frame ycf54 is required for activity of the magnesium protoporphyrin monomethylester oxidative cyclase in Synechocystis PCC 6803. J. Biol. Chem. 287: 27823-27833.
Kaneva IN, Sudbery IM, Dickman MJ, Sudbery PE (2019).Proteins that physically interact with the phosphatase Cdc14 in Candida albicans have diverse roles in the cell cycle. Sci Rep. 9(1):6258.
Cooper-Knock J, Walsh MJ, Higginbottom A, Robin Highley J, Dickman MJ, Edbauer D, Ince PG, Wharton SB, Wilson SA, Kirby J , Hautbergue GM et al (2014) Sequestration of multiple RNA recognition motif-containing proteins by C9orf72 repeat expansions.. Brain, 137(Pt 7), 2040-2051. View this article in WRRO
Wiedenheft B, van Duijn E, Bultema JB, Waghmare SP, Zhou K, Barendregt A, Westphal W, Heck AJR, Boekema EJ, Dickman MJ & Doudna JA (2011) RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. Proc Natl Acad Sci U S A, 108(25), 10092-10097.
Brouns SJJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJH, Snijders APL, Dickman MJ, Makarova KS, Koonin EV & van der Oost J (2008) Small CRISPR RNAs guide antiviral defense in prokaryotes. SCIENCE, 321(5891), 960-964. View this article in WRRO
- Protein post translational modifications
-
Cole J, Hanson EJ, James DC, Dockrell DH & Dickman MJ (2019) Comparison of data acquisition methods for the identification and quantification of histone post-translational modifications on a Q Exactive HF hybrid quadrupole Orbitrap mass spectrometer. Rapid Communications in Mass Spectrometry
Cruz-Migoni A, Hautbergue GM, Artymiuk PJ, Baker PJ, Bokori-Brown M, Chang C-T, Dickman MJ, Essex-Lopresti A, Harding SV, Mahadi NM , Marshall LE et al (2011) A Burkholderia pseudomallei toxin inhibits helicase activity of translation factor eIF4A. Science, 334(6057), 821-824.
Couto, N., Davlyatova, L., Evans, C.A. and Wright, P.C., 2018. Application of the broadband collision‐induced dissociation (bbCID) mass spectrometry approach for protein glycosylation and phosphorylation analysis. Rapid Communications in Mass Spectrometry, 32(2), pp.75-85.
Strutton B, Jaffé SRP, Pandhal J & Wright PC (2017) Producing a glycosylating Escherichia coli cell factory: The placement of the bacterial oligosaccharyl transferase pglB onto the genome. Biochemical and Biophysical Research Communications. View this article in WRRO
Evans CA, Rosser R, Waby JS, Noirel J, Lai D, Wright PC, Williams EA, Riley SA, Bury JP and Corfe BM. (2015) Reduced keratin expression in colorectal neoplasia and associated fields is reversible by both diet and resection. BMJ Gastroenterology 2: e000022
Dickman MJ, Kucharski R, Maleszka R & Hurd PJ (2013) Extensive histone post-translational modification in honey bees. Insect Biochem Mol Biol, 43(2), 125-137.
Snijders APL, Hung M-L, Wilson SA & Dickman MJ (2010) Analysis of arginine and lysine methylation utilizing peptide separations at neutral pH and electron transfer dissociation mass spectrometry.. J Am Soc Mass Spectrom, 21(1), 88-96.
Snijders APL, Pongdam S, Lambert SJ, Wood CM, Baldwin JP & Dickman MJ (2008) Characterization of post-translational modifications of the linker histories H1 and H5 from chicken erythrocytes using mass spectrometry. J Proteome Res, 7(10), 4326-4335.
- Characterisation of biotherapeutic biomolecules
-
Close ED, Nwokeoji AO, Milton D, Cook K, Hindocha DM, Hook EC, Wood H & Dickman MJ (2016) Nucleic acid separations using superficially porous silica particles. Journal of Chromatography A, 1440, 135-144. View this article in WRRO
Strutton B, Jaffé SRP, Pandhal J & Wright PC (2017) Producing a glycosylating Escherichia coli cell factory: The placement of the bacterial oligosaccharyl transferase pglB onto the genome. Biochemical and Biophysical Research Communications. View this article in WRRO
Chiverton LM, Evans C, Pandhal J, Landels AR, Rees BJ, Levison PR, Wright PC & Smales CM (2016) Quantitative definition and monitoring of the host cell protein proteome using iTRAQ - a study of an industrial mAb producing CHO-S cell line. Biotechnology Journal, 11(8), 1014-1024. View this article in WRRO
- Nucleic Acid Mass Spectrometry
-
Kung A-W, Kilby PM, Portwood DE & Dickman MJ (2018) Quantification of dsRNA using stable isotope labeling dilution liquid chromatography/mass spectrometry. Rapid Communications in Mass Spectrometry, 32(7), 590-596. View this article in WRRO
Nwokeoji AO, Kung A-W, Kilby PM, Portwood DE & Dickman MJ (2017) Purification and characterisation of dsRNA using ion pair reverse phase chromatography and mass spectrometry. Journal of Chromatography A, 1484, 14-25. View this article in WRRO
Waghmare SP, Nwokeoji AO & Dickman MJ (2015) Analysis of crRNA Using Liquid Chromatography Electrospray Ionization Mass Spectrometry (LC ESI MS), Methods Mol Biol,1311, 133-45.
Sinkunas T, Gasiunas G, Waghmare SP, Dickman MJ, Barrangou R, Horvath P & Siksnys V (2013) In vitro reconstitution of Cascade-mediated CRISPR immunity in Streptococcus thermophilus. EMBO Journal, 32(3), 385-394. View this article in WRRO
Waghmare SP & Dickman MJ (2011) Characterization and quantification of RNA post-transcriptional modifications using stable isotope labeling of RNA in conjunction with mass spectrometry analysis. Anal Chem, 83(12), 4894-4901.
Jore MM, Lundgren M, Van Duijn E, Bultema JB, Westra ER, Waghmare SP, Wiedenheft B, Pul U, Wurm R, Wagner R , Beijer MR et al (2011) Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nature Structural and Molecular Biology, 18(5), 529-536.
- Metaproteomics
-
Russo D, Beckerman AP, Couto N & Pandhal J (2016) A metaproteomic analysis of the response of a freshwater microbial community under nutrient enrichment.. Frontiers in Microbiology, 7, 1172-1172. View this article in WRRO

Experience Sheffield for yourself
The best way to find out what studying at Sheffield is like is to visit us. You'll get a feel for the atmosphere, the people, the campus and the city.
