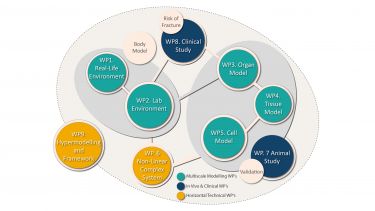
Research areas
The vision of the MultiSim programme is to develop a modelling framework, focused on the human musculoskeletal system but designed as a generic platform to address other engineering challenges that involve multi-scale modelling, unobservable states and variables, and uncertainty.
The programme is broken down into individual work packages that have their own set of objectives and desired outcomes.
As well as striving to achieve individual targets, the work packages also merge and overlap with each other.
The MultiSim Work Package Flow Chart diagram (PDF, 1.4MB) demonstrates how the work packages feed into project and affect each other – often providing a collaborative approach to problem solving and achieving. These relationships are expanded on in the individual work package descriptions.
The work packages are fall into three groups, which are described below.
Multiscale modelling
Multiscale Modelling is made up of the following five work packages (WP):
- Work Package 1. Environment Modelling
-
WP Leader: Professor Aleksandar Pavic
Main title: Medical Grade Outdoor Measurement of Human Body Dynamics.
Advancements in sensory technologies and evolution of ‘Big data’ handling techniques and infrastructure have created a unique opportunity for the healthcare sector to realise the notion of real-life data-driven diagnosis, prognosis and health monitoring to the fullest.
Rapid adoption of ‘informed’ life-style by masses in recent years has proven the strong public desire for a paradigm shift in health care services towards more personal and ‘point-of-care’ forms.
We are working to develop wearable technology for real-life continuous measurement of human body dynamics. Compared to the conventional body motion capture systems and force plates, kinematic and kinetic measurement using inertial measurement units (IMUs) and wearable force/pressure sensors offer better economy, efficiency, versatility and practicality.
Our target applications for real-life measurement are:
Estimation of spectrum of environmental boundary conditions for different activities of daily living (ADL) Musculoskeletal/neuromotor health monitoring Fall prediction and monitoring Seamless integration of wearable robots by developing ‘rich’ data-driven control technicsThe latter is mainly done using inductive statistics and models from nonlinear system identification to reveal relationships, dependencies and perform estimations of outcomes and behaviours.
The main focus of work package 1 in MultiSim project is to develop an optimised wearable sensory network combined with biomechanical and mathematical models to monitor dynamics of human body in real-life context and estimate the spectrum of ‘Environmental’ boundary conditions for different ADLs (walking, stair climbing, etc.), and pathological (tripping, falling) conditions.
The objective of this WP is to provide realistic data and environment modelling for the understanding of real physical activity of patients in their own environment and for providing input data for the body level in WP2.
Within 5C level architecture, our research is primarily focused on optimisation of sensor type and location and sensor-level data processing at ‘connection’ level, and efficient and accurate information inference at ‘conversion’ level.
Accordion content 1. - Work Package 2. Body Model
-
WP Leader: Dr Claudia Mazzà
The objective of this WP is to develop a computational framework at the body level to study the patient locomotor function through gait analysis and to define the joint forces and muscles forces to be used as input in WP3.
Standard modelling practice involves scaling a generic model, ie. the relevant bones and muscles, to a patient’s segment lengths. However, the joint and muscle forces are very sensitive to individual characteristics, (eg. bone shapes or deformities or muscle lengths and insertions), which can differ significantly from a scaled generic model.
Therefore, in the current project, patient-specific models are developed, using clinical gait analysis (CGA) and magnetic resonance imaging (MRI) data.
The problem of modelling the variables that affect the physical and physiological processes movements enabling coordination will also be tackled. In particular, the movement variability and the estimate of the muscle forces under sub-optimal neuromotor control will be estimated and used as input to the model to quantify the risk of fall and fracture.
The protocols for the gait analysis and MRI data have been defined in order to allow a full representation of gait that includes the details of the foot and ankle joints. The problem of developing a multisegment foot has also been dealt with.
This model requires the ground reaction force (GRF) to be subdivided between the modelled foot segments. A sensitivity analysis to the number of segments to be included has been performed and an available retrospective dataset combining plantar pressure data and ground reaction forces is being analysed to investigate the feasibility of using foot kinematics and morphology to predict the vertical loading that occurs in each of three foot segments during experimental walking trials.
WP2 is interacting with WP1 and WP6 for what concerns the possibility of estimating the ground reaction forces and the centre of pressure trajectory from wearable sensors. The interaction with WP6 will also concern the definition of a method that provides quantitative comparison between inverse-dynamic predicted activations and EMG measurements.
The interaction with WP3 will be based on providing them with the estimate of a spectrum of possible boundary conditions.
Last but not least, WP2 has substantially contributed to the definition of the clinical protocol to be used in the WP8 clinical study and will provide support in the data collection at whole body level.
- Work Package 3. Organ Model
-
WP Leader: Dr Xinshan Li
“Today we can to predict the strength of each bone of a given patient by simply taking a CT scan of the bone of interest”.
The Organ Model WP aims to develop modelling methods to predict changes in the musculoskeletal system at the organ scale. This typically involves the modelling of whole bones, but also of whole muscles, ligaments, etc. We mostly use finite element analysis as primary numerical method.
We started analysing the current methods for whole bone modelling. We used a retrospective cohort of 100 osteoporosis patients, half with a femoral neck fracture, provided by Prof Eastell.
We found that changes in the quality of the FE mesh and of the anatomical reference system (used to define the musculo-articular forces acting on the femur), significantly improve the ability of the minimal physiological strength to discriminate fractured and non fractured patients.
As one of the main sources of error was found to be the CT scan, that was limited to the proximal femur, we developed a new CT scan plan that cover the entire femur, but present an effective radiation dose smaller than the current clinical protocol.
We are also working on modelling paediatric fractures in the long bones (femurs and tibia), and on the and on the modelling of skeletal muscles.
We will soon start to receive skeletal forces to be applied to our models from WP2, as these are predicted by the whole body models.
From WP4 we would expect predictions of the evolution of the apparent density over time due to the progression of the disease (osteoporosis, metastasis, ostemalachia), or as an effect of the intervention.
- Work Package 4. Tissue Modelling
-
WP Leader: Dr Enrico Dall’Ara
The objective of this WP is to develop mechanoregulation models able to predict the tissue formation and resorption as a function of the mechanical loading.
In particular bone topological discretisation and bone remodelling algorithms used in combination with mechanoregulation models will be developed. Also a new constitutive model of muscles will be developed.
These models will be integrated into the Organ Modelling WP3 and will make use of the outputs provided by the Cell Modelling WP5.
Volumetric Topological Analysis (VTA) is considered as a powerful complex geometry analysis scheme which provides important information about trabecular network architects and its contribution to whole musculoskeletal mechanical response.
Two methodologies were applied in this study. In the first phase, an algorithm was developed in MATLAB to perform medial surface thinning on a trabecular microstructure. In the second phase, a separate MATLAB program was generated to perform digital topological analysis which aims to organise thinned trabecular bone network into separated plate and rod members.
Therefore, by applying our VTA package on any trabecular microstructure, it is then possible to extract detailed information on structural topology, network formation and local anisotropy. This technique can also be used to obtain more knowledge on the effect of diseases, such as osteoporosis, over the bone microarchitecture.
The topological architecture developed in this WP will provide inputs in terms of anisotropic and stiffness properties of bone at the organ level (WP5).
- Work Package 5. Cell Model
-
WP Leader: Professor Tim Skerry
The objective of this WP is to gain knowledge on the mechanoresponse of cells when subjected to mechanical loading and to integrate single cell level and population-based level experimental and numerical information into the tissue scale WP. I
n particular a dynamic single cell computational model simulating the cytoskeleton adaptation of cell to mechanical loading will be developed.
Cell-cell interactions will then be studied by combining agent-based modelling technique with the single cell finite element model. This will provide a new approach to model cells individually or in a population.
The output of this approach will enable to study in vitro processes that will inform on the tissue extracellular matrix formation.
The types of mechanical loading will be derived from the outputs provided in WP4. In addition some modelling at the single cell level and population-based level will be performed in order to integrate it with the tissue WP4.
On the other side the mechanotransduction of osteocyte cells will be studied experimentally by taking RNA sequence of cells in the skull and long bones to evaluate the effect of in vivo mechanical loading onto gene expression patterns.
The outcome of these experimental mechanical stimulations on osteocytes transcriptomic response will likely contribute to the identification of mediators involved in the adaptive response of bone.
Cell memory and site-specific response will also be studied by investigating the effect of mechanical loading in vitro on osteocyte cells extracted from the tibia and skull of different animals.
Cross Cutting
- Work Package 6. Non-linear complex systems
-
WP Leader: Dr Lingzhong Guo
Co-Leader: Professor Visakan Kadirkamanathan
The objective of this WP is to develop new approaches to model non-linear systems that have uncertain and incomplete information through unobservable states and variables.
The approaches will be based on the NARMAX methodology. Acting as a horizontal WP, this WP aims to integrate these approaches within the musculoskeletal models developed in WP1-5 and to provide support for all Work Packages relating to nonlinear signal processing, system identification, modelling, and analysis.
Output to date: We are working on four main themes: proxy measurements, model reduction, multiscale linking, and nonlinear system identification and signal processing.
This involves the derivation of new algorithms to address each of these problems from a generic systems engineering perspective with applications to solving specific problems that have arisen from the other MultiSim Work Packages.
In each case we have derived new solutions to these problems and have completed feasibility studies on specific applications from the other MultiSim Work Packages with excellent results. WP6 interacts with all of the other Work Packages.
In Vivo and Clinical
In Vivo and Clinical consists of two work packages:
- Work Package 7. Animal Study
-
WP Leader: Dr Enrico Dall’Ara
“Animal studies will allow us to challenge the prediction ability of our computation models in each subject in vivo, and therefore to improve our understanding of the link between cell, tissue and organ levels”
Animal models can be used to obtain missing information not available in humans and to quantitatively validate the computational models.
The models created in this WP will be based on the ones developed in the other work packages and adapted for mice bones, in particular for the tissue and organ levels.
The developed models based on state of the art imaging techniques will be challenged in predicting induced bone resorption and bone apposition.
Ovariectomy or castration procedures will be used in order to study local and systemic regulators of bone resorption and morphological changes. Mice will be allowed to lose bone for 2-4 weeks.
To model physiological increases in bone mass, non-invasive application of mechanical loads to the tibia will be performed on alternate days in a 2-week period. Drug treatments to moderate these effects will include bisphosphonates (to inhibit bone loss) and PTH to stimulate bone accrual.
The WP7 has started only in the second year of the project and, therefore, the preliminary data are mainly related to the work done in parallel projects. An in vivo microCT scanning procedure has been developed in order to scan the mouse tibia of the same mouse longitudinally.
In approximately 30 minutes time we can scan the whole tibia and fibula with high resolution (10 microns voxel size) and measure trabecular and cortical densitometric and morphometric parameters. The reduction of scanning time and radiation dose will be investigated during the next months.
A loading model for the mice tibia has been developed in Prof Tim Skerry’s group where both tibia and fibula are loaded repeatedly. From preliminary data bone apposition has been seen by means of microCT ex vivo. Now we are replicating this approach to track the changes in the same bone of the sample animal in vivo.
With WP4 (tissue level). The procedures currently defined in WP4 for developing models of cortical and trabecular bone for human tissue are used in order to create models at the organ level for the mice bones.
In particular, similar microFE approach will be used in order to convert high resolution microCT images into large hexahedral mesh models. With WP5 (cell level).
The experimental part of WP7 will be integrated to the analysis of the RNA in order to explore different gene expression in loaded and not loaded bones.
- Work Package 8. Clinical Study
-
WP Leader: Professor Eugene McCloskey
The objective of this WP is to gather clinical data for the WP1-5 and to test the multiscale hypermodelling framework within a clinical context.
A number of subjects (including fallers and non-fallers) will be recruited to form a complete database of potential clinical imaging and functional assessments that will be made available for all WPs.
For these subjects gait analysis (WP2) will be performed in the lab while sensors in their home developed in WP1 will be used for a short period of time (1 week). QCT and MRI of the lower limb will be obtained to obtain bone, cartilage, ligament and muscle information (geometry and pixel-intensity related information).
Other
- Work Package 9. Hypermodelling integration and framework development
-
WP Leader: Dr Pinaki Bhattacharya
The objective of this work package is to build a multiscale framework to link the models at different scales: WP1 to WP5.

MultiSim Project
Aiming to create a new generation of predictive models capable of handling complex multi-scale and multiphysics problems, characterised by uncertain and incomplete information.