COVID-19
University of Sheffield scientists are contributing to the fight against coronavirus.
After sequencing the whole-genomes of SARS-CoV-2 from two local COVID-19 positive patient samples1, University of Sheffield researchers are now involved in generating and sharing sequence data as part of the COVID-19 Genomics UK (COG-UK) – Consortium2.
COG-UK Consortium
The Government, NHS, Public Health England, UK Research and Innovation and the Wellcome Trust are collectively investing £20 million into sequencing the UK SARS-CoV-2 epidemic at institutions across the country2.
This money will drive the sequencing capabilities of Public Health England, Wellcome Sanger Institute and centres in Belfast, Birmingham, Cambridge, Cardiff, Edinburgh, Exeter, Glasgow, Liverpool, London, Norwich, Nottingham, Oxford and Sheffield.
At the University of Sheffield, the ARTIC pipeline will be used to sequence the SARs-CoV-2 virus, using protocols and bioinformatics based on the Oxford Nanopore Technologies fourth-generation sequencers3.
COG-UK expect to generate thousands of SARS-CoV-2 genomes per week and the resulting genomic data will be used to map out the epidemiology and phylogeny of the virus.
The results from sequencing across UK centres will be condensed and reported to the government weekly, to provide information on virus progression and the potential impact of public health measures to limit the spread of SARS-CoV-2 in the UK.
These enhanced capabilities will remain in place for future research and pandemic preparedness.
The University of Sheffield team
Management
- Thushan de Silva, Senior Clinical Lecturer and Honorary Consultant Physician in Infectious Diseases from the University of Sheffield’s School of Medicine and Population Health, is leading a team of individuals from across IICD, the Sheffield Institute for Translational Neuroscience (SITraN), The Department of Computer Science and Molecular Biology and Biotechnology (MBB), including the Department of Animal and Plant Sciences (APS).
- Matthew Parker from SITraN and the University of Sheffield's Bioinformatics Core is leading the data organisation and analysis, locally.
- Nikki Smith from the School of Medicine and Population Health is acting as project manager for the Sheffield COG-UK team, supported by Katie Cooke from the school's technician team.
Diagnostics
- Consultant Virologists Cariad Evans, Mohammad Raza and Alison Cope are leading the testing of COVID-19 patients and staff at the Sheffield Teaching Hospitals (STH) NHS Foundation Trust Virology Department, and collaborating on this project.
- Alex Keeley, an Academic Clinical Fellow in Infectious Diseases and Dave Partridge, Consultant Microbiologist, are coordinating the use of patient samples for sequencing, managing the corresponding clinical data, along with the STH IT department.
Sequencing and Analysis
- Sheffield Institute of Translational Neuroscience/Sheffield Biomedical Research Centre: Matthew Wyles oversees the running of the UoS Next Generation Sequencing core facility, and has facilitated the upscaling of sequencing capabilities in the School of Medicine and Population Health. In addition to Matthew Parker and Dennis Wang (Department of Computer Science and School of Medicine and Population Health), Bioinformatics core director Mark Dunning and PhD researchers Sokratis Karitois and Katjusa Koler, have helped to develop the bioinformatics pipelines and interfaces which will be used by the University of Sheffield.
- School of Medicine and Population Health: Post-Doctoral Research Associates Adri Angyal, Rebecca Brown and Luke Green have been leading the laboratory work, along with research technicians Danielle Groves and Hailey Hornsby. Ben Lindsey, an Academic Clinical Fellow in Infectious Diseases and Virology is helping with local phylogenetic analysis.
- APS: Senior Research Technicians Rachel Tucker and Paul Parsons have kindly joined the team after the offer of help from Prof. Terry Burke, and are helping to scale up the process to generate data for up to 230 genomes per week.
IT Support
- IT Services: IT services are providing infrastructure and high performance computing capabilities which enable analysis for the project to be performed.
- Department of Computer Science: Computer science have provided IT equipment and expertise through Dennis Wang to allow researchers on the project complete the complex analysis required to produce meaningful results from the viral sequencing data.
How does it work?
Patient samples are taken, most commonly from the nose or throat, before they are purified and the RNA is amplified to identify the genes unique to coronavirus4.
The COVID-19 genome is encoded by RNA and is approximately 30,000 bases in length. RNA samples are converted to complementary DNA (cDNA) using a reverse transcriptase enzyme. cDNA is amplified into short overlapping DNA fragments repeatedly by polymerase chain reaction (PCR)5. Genome sequencing requires many copies of DNA to produce many iterations of the genome. The number of times a particular area of the genome has been sequenced is known as the ‘coverage’, and the greater the coverage the more confidence we have in the output.
Up to 23 genomes can be sequenced in one run by adding specific barcodes to the overlapping DNA sequences. Adaptor sequences are also added to ensure the DNA passes through the pores of the MinION sequencer (https://nanoporetech.com/how-it-works). As the DNA passes through the pore this causes a disruption in the electric current running through the solution, the resulting electrograms can be converted by an algorithm to give the sequence of nucleic acids5. The sequences are separated by barcode and aligned to previously sequenced COVID-19 sequences using Read Assignment, Mapping, and Phylogenetic Analysis in Real Time (RAMPART)6.
The sequence data generated will be uploaded to https://www.gisaid.org/ and other public data repositories for sharing with the wider scientific community7.
Big Picture
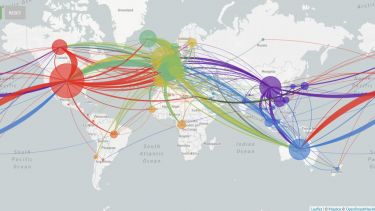
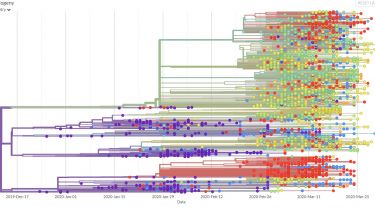
Viral genomes are constantly accumulating mutations and evolving. Tracking these changes can build a picture of the spread of the virus, identifying transmission chains, including estimates of asymptomatic spread8.
This information could be used retrospectively to identify how the SARs-CoV-2 virus entered the human population and its subsequent evolution. The phylogeny of the virus is being mapped out on https://nextstrain.org/ncov9 and http://virological.org/10 and could be used to identify evolutionary drivers, such as the impact of public health interventions, natural immunity or treatment interventions11.
References
- Whole genomes of coronavirus from UK patients sequenced by Sheffield scientists - Latest - News - The University of Sheffield. Available at: https://www.sheffield.ac.uk/news/nr/coronavirus-genomes-sequenced-sheffield-1.883916. (Accessed: 2nd April 2020)
- UK launches whole genome sequence alliance to map spread of coronavirus - GOV.UK. Available at: https://www.gov.uk/government/news/uk-launches-whole-genome-sequence-alliance-to-map-spread-of-coronavirus. (Accessed: 2nd April 2020)
- Artic Network. Available at: https://artic.network/ncov-2019/ncov2019-bioinformatics-sop.html. (Accessed: 1st April 2020)
- WHO. World Health Organization. Laboratory testing for coronavirus disease 2019 (COVID-19) in suspected human cases. Interim Guid. 1–7 (2020). https://apps.who.int/iris/bitstream/handle/10665/331329/WHO-COVID-19-laboratory-2020.4-eng.pdf?sequence=1&isAllowed=y
- Quick, J., Barnes, K. & Quick, J. nCoV-2019 sequencing protocol. 1–24 (2020).
- GitHub - artic-network/rampart: Read Assignment, Mapping, and Phylogenetic Analysis in Real Time. Available at: https://github.com/artic-network/rampart. (Accessed: 1st April 2020)
- Shu, Y. & McCauley, J. GISAID: Global initiative on sharing all influenza data – from vision to reality. Eurosurveillance 22, (2017). https://www.eurosurveillance.org/content/10.2807/1560-7917.ES.2017.22.13.30494
- Grubaugh, N. D. et al. Tracking virus outbreaks in the twenty-first century. Nature Microbiology 4, 10–19 (2019). https://www.nature.com/articles/s41564-018-0296-2
- Hadfield, J. et al. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics 34, 4121–4123 (2018). https://academic.oup.com/bioinformatics/article/34/23/4121/5001388
- Phylodynamic Analysis | 176 genomes | 6 Mar 2020 - Novel 2019 coronavirus / nCoV-2019 Genomic Epidemiology - Virological. Available at: http://virological.org/t/phylodynamic-analysis-176-genomes-6-mar-2020/356. (Accessed: 1st April 2020)
- Grenfell, B. T. et al. Unifying the Epidemiological and Evolutionary Dynamics of Pathogens. Science 303, 327–332 (2004). https://science.sciencemag.org/content/303/5656/327
Latest news
Publications
- Starting to understand the biology of Covid-19
-
The current COVID-19 pandemic is transforming the way that science is being undertaken with novel collaborative groupings being established like never before, sharing expertise, time and data in an effort to understand the biology of the virus.
Professor Colin Bingle, from the School of Medicine and Population Health has been part of one such global collaborative project to understand the specific cell types which are targeted by SARS-CoV-2 (the virus that causes COVID-19 disease) in the respiratory tract. This work, that was recently published in Cell (10.1016/j.cell.2020.04.035), stemmed from an existing collaboration with Dr Jose Ordovas-Montanes at Boston Children’s Hospital looking at gene expression in specific cell populations in the upper respiratory tract. For this project Jose and Dr Alex Shalek, from Massachusetts Institute of Technology and Harvard remotely brought together groups from across the world, including from the Human Cell Atlas Lung Biological Network, to study the cell types that express the key viral receptor protein angiotensin-converting enzyme 2 (ACE2). Normally, this cell surface enzyme plays a role in helping the body maintain blood pressure and fluid balance. Very early in the pandemic it was shown that ACE2 and a cell surface protease TMPRSS2 were needed to generate a functional receptor pathway in cells targeted by the virus.
This study investigated a wealth of independent data sets generated using single cell RNA sequencing (scRNAseq) to identify the specific cell types that co-express ACE2 and TMPRSS2. This showed an enrichment in different cell types in different locations. In the nasal passages, it was mucus-producing goblet secretory cells; in the lung it was alveolar type II pneumocytes and in the intestine, it was the enterocytes. Computational analysis suggested that elevation of ACE2 expression might be associated with an interferon response, a protein that the body makes in response to viral infections and this was confirmed by treatment of airway cells with interferon. Importantly, this analysis also showed that this response was not seen in mice. The main conclusion from the paper is the suggestion that SARS-CoV-2 (and potentially other coronaviruses) that use ACE2 as a receptor for infection may have evolved take advantage of the body’s natural defences. When the body responds to the infection by producing more interferon, that in turn results in production of more ACE2, enhancing the ability of the virus to attach more readily to lung cells. The true consequence of this, like so much SARS-CoV2 biology, remains to be fully understood.
- Periscope: sub-genomic RNA identification in SARS-CoV-2 ARTIC Network Nanopore Sequencing Data
-
We have developed periscope, a tool for the detection and quantification of sub-genomic RNA in ARTIC network protocol generated Nanopore SARS-CoV-2 sequence data. We applied periscope to 1155 SARS-CoV-2 sequences from Sheffield, UK.
Using a simple local alignment to detect reads which contain the leader sequence we were able to identify and quantify reads arising from canonical and non-canonical sub-genomic RNA.
We were able to detect all canonical sub-genomic RNAs at expected abundances, with the exception of ORF10, suggesting that this is not a functional ORF. A number of recurrent non-canonical sub-genomic RNAs are detected.
We show that the results are reproducible using technical replicates and determine the optimum number of reads for sub-genomic RNA analysis.
Finally variants found in genomic RNA are transmitted to sub-genomic RNAs with high fidelity in most cases.
This tool can be applied to tens of thousands of sequences worldwide to provide the most comprehensive analysis of SARS-CoV-2 sub-genomic RNA to date. doi: https://www.biorxiv.org/content/10.1101/2020.07.01.181867
- Tracking changes in SARS-CoV-2 Spike: evidence that D614G increases infectivity of the COVID-19 virus
-
A SARS-CoV-2 variant carrying the Spike protein amino acid change D614G has become the most prevalent form in the global pandemic. Dynamic tracking of variant frequencies revealed a recurrent pattern of G614 increase at multiple geographic levels: national, regional and municipal. The shift occurred even in local epidemics where the original D614 form was well established prior to the introduction of the G614 variant. The consistency of this pattern was highly statistically significant, suggesting that the G614 variant may have a fitness advantage. We found that the G614 variant grows to higher titer as pseudotyped virions. In infected individuals G614 is associated with lower RT-PCR cycle thresholds, suggestive of higher upper respiratory tract viral loads, although not with increased disease severity. These findings illuminate changes important for a mechanistic understanding of the virus, and support continuing surveillance of Spike mutations to aid in the development of immunological interventions.
- Ethics of controlled human infection to address COVID-19
-
Tom Darton, Florey Advanced Clinical Fellow in the School of Medicine and Population Health has recently published a paper in Science (doi: 10.1126/science.abc1076) looking at the ethics of human challenge studies to accelerate the development of a vaccine to prevent COVID-19 in the current pandemic.
While social distancing has had a major impact in reducing the current scale of the UK pandemic, widespread us of a safe and effective vaccine is seen by many as the only way of returning to a normal way of life. To accelerate the vaccine development pipeline, human challenge studies are often used to provide vital information regarding the human immune response to infection or whether new vaccine candidates are likely to work in preventing infection. In these studies, healthy human volunteers are deliberately exposed to infection, and so minimising the risks to study participants, researchers and the community is vital.
Evaluating the risks and benefits of any challenge study is highly complex, and this calculation changes still further in the context of a global pandemic. While the risks may be higher due to, for example, the lack of available rescue treatments, the benefits to society in accelerating vaccine availability even by a couple of weeks could save many lives. The availability of a human challenge study becomes particularly relevant when the number of cases (or “R0) falls. While this is good news, it makes vaccine evaluation more difficult and protracts the timeline. With an anticipated second wave anticipated, using a challenge study to expedite vaccine evaluation and/or to prioritise selection before the numbers of cases occurring in the community starts to increase could have a major impact.
Paper conclusion: With increasing interest in the ethical issues raised by human challenge studies, there is now broad consensus on many of the key issues including acceptability of risk, the meaning of informed consent, and proportionate compensation to participants. In a pandemic situation, however, the social value of this research becomes a more significant factor which may, or may not, justify a higher level of risk and uncertainty. Here we provide a ethical framework to support discussions around the development of COVID-19 human challenge studies.
- Roll-out of SARS-CoV-2 testing for healthcare workers at a large NHS Foundation Trust in the United Kingdom, March 2020
-
Healthcare workers (HCW) are potentially at increased risk of infection with coronavirus disease (COVID-19) and may transmit severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) to vulnerable patients. We present results from staff testing at Sheffield Teaching Hospitals NHS Foundation Trust, United Kingdom. Between 16 and 29 March 2020, 1,533 symptomatic HCW were tested, of whom 282 (18%) were positive for SARS-CoV-2. Testing HCW is a crucial strategy to optimise staffing levels during this outbreak. doi: 10.2807/1560-7917.ES.2020.25.14.2000433
- Evaluating use cases for human challenge trials in accelerating SARS-CoV-2 vaccine development
-
Human challenge trials (HCTs) have been proposed as a means to accelerate SARS-CoV-2 vaccine development. We identify and discuss three potential use cases of HCTs in the current pandemic: evaluating efficacy, converging on correlates of protection, and improving understanding of pathogenesis and the human immune response. We outline the limitations of HCTs and find that HCTs are likely to be most useful for vaccine candidates currently in preclinical stages of development. We conclude that, while currently limited in their application, there are scenarios in which HCTs would be extremely beneficial. Therefore, the option of conducting HCTs to accelerate SARS-CoV-2 vaccine development should be preserved. As HCTs require many months of preparation, we recommend an immediate effort to (1) establish guidelines for HCTs for COVID-19; (2) take the first steps toward HCTs, including preparing challenge virus and making preliminary logistical arrangements; and (3) commit to periodically re-evaluating the utility of HCTs. doi: 10.1093/cid/ciaa935
- Broad and strong memory CD4⁺ and CD8⁺ T cells induced by SARS-CoV-2 in UK convalescent COVID-19 patients
-
COVID-19 is an ongoing global crisis in which the development of effective vaccines and therapeutics will depend critically on understanding the natural immunity to the virus, including the role of SARS-CoV-2-specific T cells. We have conducted a study of 42 patients following recovery from COVID-19, including 28 mild and 14 severe cases, comparing their T cell responses to those of 16 control donors. We assessed the immune memory of T cell responses using IFNγ based assays with overlapping peptides spanning SARS-CoV-2 apart from ORF1. We found the breadth, magnitude and frequency of memory T cell responses from COVID-19 were significantly higher in severe compared to mild COVID-19 cases, and this effect was most marked in response to spike, membrane, and ORF3a proteins.
Total and spike-specific T cell responses correlated with the anti-Spike, anti-Receptor Binding Domain (RBD) as well as anti-Nucleoprotein (NP) endpoint antibody titre (p<0.001, <0.001 and =0.002). We identified 39 separate peptides containing CD4⁺ and/or CD8⁺ epitopes, which strikingly included six immunodominant epitope clusters targeted by T cells in many donors, including 3 clusters in spike (recognised by 29%, 24%, 18% donors), two in the membrane protein (M, 32%, 47%) and one in the nucleoprotein (Np, 35%). CD8⁺ responses were further defined for their HLA restriction, including B*4001-restricted T cells showing central memory and effector memory phenotype. In mild cases, higher frequencies of multi-cytokine producing M- and NP-specific CD8⁺ T cells than spike-specific CD8⁺ T cells were observed. They furthermore showed a higher ratio of SARS-CoV-2-specific CD8⁺ to CD4⁺ T cell responses. Immunodominant epitope clusters and peptides containing T cell epitopes identified in this study will provide critical tools to study the role of virus-specific T cells in control and resolution of SARS-CoV-2 infections.
The identification of T cell specificity and functionality associated with milder disease, highlights the potential importance of including non-spike proteins within future COVID-19 vaccine design. doi: 10.1101/2020.06.05.134551
- An integrated national scale SARS-CoV-2 genomic surveillance network
-
The Coronavirus Disease 2019 (COVID-19) Genomics UK Consortium (COG-UK) was launched in March, 2020, with £20 million support from UK Research and Innovation, the UK Department of Health and Social Care, and Wellcome Trust. The goal of this consortium is to sequence severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) for up to 230 000 patients, health-care workers, and other essential workers in the UK with COVID-19, which will help to enable the tracking of SARS-CoV-2 transmission, identify viral mutations, and integrate with health data to assess how the viral genome interacts with cofactors and consequences of COVID-19. doi: 10.1016/ S2666-5247(20)30054-9
- Asymptomatic SARS-CoV-2 infection: the tip or the iceberg?
-
We describe an outbreak on a cruise ship departing Argentina for the Antarctic peninsula with 223 members, all of whom were screened with temperature and symptom check prior to embarkation in mid-March 2020. At day 8, the first member developed symptoms. Despite immediate imposition of cabin isolation of all passengers and strict personal protective equipment use by crew, 128 (59.0%) of those on board tested positive for SARS-CoV-2 on day 20, when everyone was screened with an RT-PCR-based test. Of those testing positive, only 24 (18.8%) developed symptoms and 104 (81.2%) remained asymptomatic until day 28, when the first cohort of passengers disembarked. Some caution is required in interpreting this figure as the follow-up period is not long enough to be confident that all individuals remained truly asymptomatic. Nevertheless, it is striking that despite rigorous attempts to limit spread, such widespread transmission occurred on board. The enclosed environment may be the major driver of the high transmission rate; however, the possibility that transmission from asymptomatic cases contributed significantly must also be considered. doi: 10.1136/thoraxjnl-2020-215337
- Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK.
-
A safe and efficacious vaccine against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), if deployed with high coverage, could contribute to the control of the COVID-19 pandemic. We evaluated the safety and efficacy of the ChAdOx1 nCoV-19 vaccine in a pooled interim analysis of four trials. doi: 10.1016/S0140-6736(20)32661-1