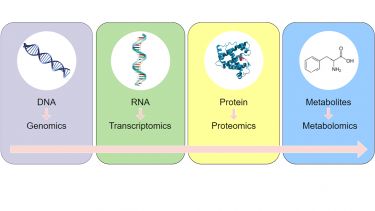
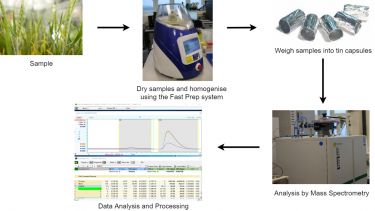
Metabolomics strategies
The biOMICS Facility provides a full infrastructure and applies diverse strategies for metabolite identification, characterization and quantification using mass spectrometry based approaches. Please do not hesitate to contact us to discuss your requirements and provide further information.
- Definition of Metabolomics/Metabolite Profiling
-
Metabolism is an integrated network of pathways where what happens in one pathway in turn affects other pathways.
Metabolomics is the comprehensive analysis of all metabolites, the by-products of metabolism. It is a high-throughput analytical strategy which gives a snapshot of which compounds are present at relative levels at a specific time point by analysing as many metabolites as possible in one screen.
However as adjacent cells in a tissue can have very different metabolic profiles and since metabolites come from many different classes of chemicals no one analytical approach can detect all components of metabolism.
Below are some of the most common strategies for metabolomics which we specialise in within the Facility.
- Untargeted Metabolite Profiling
-
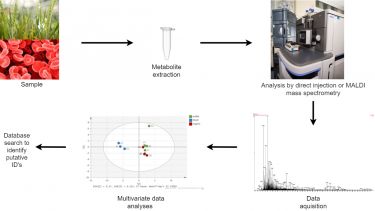
Metabolite profiling involves an un-biased, non-targeted analysis approach to identify as many metabolites as possible in a particular sample. We can target both positively and negatively charged ions.
A common approach to do this is to initially screen sample extracts for as wide a range of masses as possible using the state-of-the-art mass spectrometers available in the Facility. This is usually done using a TOF mass spectrometer which gives a wide dynamic range over a large mass range. This gives us large amounts of data which we interpret using multivariate data analysis techniques such as PCA and OPLS-DA. This is a rapid and high throughput screening method which can identify potential changing metabolites within our samples. This method does not however identify any particular compounds it merely identifies masses which are changing leading onto more targeted analyses. Database searches are used here to give potential or putative ID’s to any targets identified.
- Targeted metabolite analysis
-
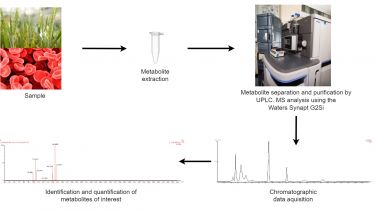
From an untargeted metabolite screening approach any masses identified as changing or different between sample groups can then be identified and quantified by more precise methods. Targeted analyses can involve using more optimised extraction methods for particular classes of compounds. They could also involve more specific separation techniques using standard UPLC or HPLC methods or more specialised convergence chromatography, or detection techniques using UV, RI or ECD detections or by using ion mobility to separate isomers or even GC-MS. The method chosen is dictated either by the experiment or from a previous untargeted analysis.
Tandem mass spectrometry is used for confirmation of the identity of any metabolites.
So for example a metabolite screen experiment could involve a non-targeted study which then indicates changes in a particular pathway. The metabolites of interest can then be positively identified and quantified using a more targeted approach.
- Mass Spectrometry Imaging
-
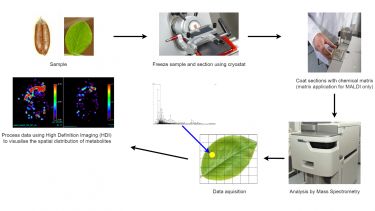
Mass spectrometry imaging (MSI) is a technique used to visualise the spatial distribution of chemical compositions e.g. compounds, biomarkers, metabolites, peptides or proteins by their molecular masses. There are several advantages to imaging. It enables the study of metabolites in situ and negates the need for complex sample preparation, which could alter metabolism. It has a high spatial resolution of (1–<30 μm), no target specific reagents are needed so reagent costs for the analysis are exceedingly low, while analysis time is exceedingly fast.
Both matrix-assisted laser desorption/ionisation mass spectrometric imaging (MALDI) and desorption electrospray ionisation mass spectrometric imaging (DESI) are analytical techniques offered by the metabolomics facility.
The diagram below shows a typical MALDI imaging workflow.
- Mobile Mass Spectrometry
-
As detailed in the instruments section we have two mobile mass spectrometers in the metabolomics facility. Both GC-MS and LC-MS systems are available. These systems have simple sample introduction methods and are capable of providing rapid analysis and screening thus improving workflow in the field. Benefits to working in the field include samples being analysed immediately (limiting changes in metabolism), no transportation and reduction in sample preparation and handling.
The photo below shows us out in the field with a mobile mass spectrometer at RAGT seeds in Cambridgeshire measuring metabolite markers in a field of wheat.
- Stable isotope analysis
-
In isotope ratio mass spectrometry, element isotope ratios are determined very accurately and precisely. Typically, single focusing magnetic sector mass spectrometers with fixed multiple detectors (one per isotope) are used. Complex compounds are reduced to simple molecules prior to measurement; for example, organic compounds are combusted to CO2, H2O and N2.
The technique involves the coupling of a preparation system employing the Dumas principle with a stable isotope mass spectrometer. This allows the measurement of not only total nitrogen or carbon in a sample but also their 15N or 13C levels in a wide range of biological and chemical samples.