Professor David Strutt
School of Biosciences
Professor of Developmental Genetics
Wellcome Trust Senior Fellow in Basic Biomedical Science


Full contact details
School of Biosciences
Firth Court
Western Bank
Sheffield
S10 2TN
- Profile
-
- 2005: Professor of Developmental Genetics
- 2003-present: Wellcome Senior Fellow in Basic Biomedical Science, University of Sheffield
- 1998-2003: Lister Institute-Jenner Research Fellow, University of Sheffield
- 1997-1998: Lecturer, University of Sheffield
- 1993-1997: Postdoctoral Fellow, European Molecular Biology Laboratory, Heidelberg
- 1991-1993: Postdoctoral Fellow, Department of Anatomy, University of Cambridge
- 1988-1991: PhD, Department of Anatomy, University of Cambridge
- 1985-1988: BA Natural Sciences, University of Cambridge
- Research interests
-
We are interested in the genetic control of animal development and how cells interact to build complex tissues and organs. We focus on the particular problem of how cells coordinate their polarity within developing tissues, and as a model study the phenomenon of planar polarity in epithelia of the fruitfly Drosophila.
Understanding mechanisms of coordinated cell polarisation during animal development
Lay summary of research
We are interested in the process of morphogenesis, which is the process by which a body grows and develops to form complex tissues and organ systems. This involves the regulation and coordination of three processes: first, the multiplication of cells; second, the differentiation of the right types of cells in the right positions; third, the correct orientation of cells relative to each other. It is this final process in which we are primarily interested.
As the orientation of cells relative to each other is a fundamental conserved process in animal development, we are able to study it using model experimental systems, in particular the fruitfly Drosophila melanogaster. Using this organism we can easily manipulate the function of genes, which has allowed the identification of a large number of genes that are involved in the coordination of cell orientation.
The aims of our research are to understand how these genes work together and how they are organised into hierarchies. By understanding how the genes act, we hope to be able to intervene to correct developmental diseases caused by dysfunction of the genes, and also gain insights into conditions such as cancer metastasis which involve coordinated changes in cell polarity.
Scientific summary of research
Morphogenesis – the formation of multidimensional structures from cells and groups of cells – is one of the fundamental processes underlying the development of complex multicellular organisms. A key property underlying many morphogenetic processes is the ability of cells to coordinate their polarity within a sheet of cells, a phenomenon referred to as "planar polarity" or "planar cell polarity [PCP]". Planar polarity enables the concerted cell movements and rearrangements necessary to shape tissues, and allows the production of arrays of polarised structures such as hairs and cilia.
Work in the Strutt lab seeks to understand molecular mechanisms of planar polarisation. Our approach is to study two conserved mechanisms, the "core" Frizzled-dependent planar polarity pathway and the Fat/Dachsous pathway. Our goal is to dissect the feedback and cell-cell communication mechanisms that mediate coordinated cellular symmetry breaking, and ask how this can be aligned with the axes of tissues.
We exploit the traditional strengths of molecular genetic analysis in Drosophila to precisely manipulate gene and protein function in vivo, taking advantage of the minimal genetic redundancy and the easy accessibility of developing tissues. In particular, this system allows us to perform in vivo cell biology, analysing protein dynamics and behaviour in the context of developing tissues.
To do this, we employ both conventional high resolution in vivo imaging techniques such as confocal microscopy, methodologies such as FRAP (fluorescence recovery after photobleaching) and also super-resolution microscopy. These in vivo studies of protein behaviour are combined with studies of the biochemical interactions of pathway components, targeted genetic screens to identify novel regulatory factors, and computational modelling.
Our long term goal is a fundamental understanding of how coordinated cell polarisation is achieved through the integrated spatiotemporal interactions of pathway components acting from the molecular (nanometer) to cellular (micrometre) to tissue (millimetre) scales.
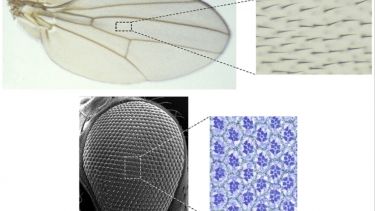
Examples of planar polarised structures on the adult cuticle of the fruitfly Drosophila. Top: photomicrograph of an adult wing (left) with inset (right) showing the regular array of planar polarised hairs on the surface of the wing. Bottom: Scanning electron micrograph of an adult eye (left) with inset (right) showing a histologically stained microtome section through the eye revealing the regular planar polarised arrangements of the groups of cells within the eye that form each facet.
- Publications
-
Show: Featured publications All publications
Featured publications
Journal articles
- Molecular symmetry breaking in the Frizzled-dependent planar polarity pathway. Current Biology, 33(24), 5340-5354.e6. View this article in WRRO


- Distinct mechanisms of planar polarization by the core and Fat-Dachsous planar polarity pathways in the Drosophila wing. Cell Reports, 40(13).


- QuantifyPolarity, a new tool-kit for measuring planar polarized protein distributions and cell properties in developing tissues. Development, 148(18). View this article in WRRO


- Reciprocal action of casein kinase Iε on core planar polarity proteins regulates clustering and asymmetric localisation. eLife, 2019(8).


- Retromer controls planar polarity protein levels and asymmetric localization at intercellular junctions. Current Biology, 29(3), 484-491.e6. View this article in WRRO


- Rapid disruption of dishevelled activity uncovers an intercellular role in maintenance of prickle in core planar polarity protein complexes. Cell Reports, 25(6), 1415-1424.e6. View this article in WRRO


- A Dual Function for Prickle in Regulating Frizzled Stability during Feedback-Dependent Amplification of Planar Polarity. Current Biology, 27(18), 2784-2797.e3. View this article in WRRO


- Robust Asymmetric Localization of Planar Polarity Proteins Is Associated with Organization into Signalosome-like Domains of Variable Stoichiometry.. Cell Rep, 17(10), 2660-2671.


- Dynamics of core planar polarity protein turnover and stable assembly into discrete membrane subdomains.. Dev Cell, 20(4), 511-525.


All publications
Journal articles
- Adhesion G protein-coupled receptors. Pharmacological Reviews, 100116-100116.


- Characterisation of cell-scale signalling by the core planar polarity pathway during Drosophila wing development.


- Tissue shear as a cue for aligning planar polarity in the developing Drosophila wing. Nature Communications, 16(1). View this article in WRRO


- The Fat-Dachsous planar polarity pathway competes with hinge contraction to orient polarized cell behaviors during Drosophila wing morphogenesis. Current Biology, 35(2), 422-430.e3. View this article in WRRO


- Fat-Dachsous planar polarity function requires two distinct heterophilic cadherin-cadherin binding interactions. Cell Reports, 43(10), 114722-114722.


- Recombinant biosensors for multiplex and super-resolution imaging of phosphoinositides. Journal of Cell Biology, 223(6). View this article in WRRO


- Geometry-preserving expansion microscopy microplates enable high-fidelity nanoscale distortion mapping. Cell Reports Physical Science, 4(12). View this article in WRRO


- Molecular symmetry breaking in the Frizzled-dependent planar polarity pathway. Current Biology, 33(24), 5340-5354.e6. View this article in WRRO


- Deciphering the roles of cell shape and Fat and Dachsous planar polarity in arranging the Drosophila apical microtubule network through quantitative image analysis. Molecular Biology of the Cell.


- Distinct mechanisms of planar polarization by the core and Fat-Dachsous planar polarity pathways in the Drosophila wing. Cell Reports, 40(13).


- Selective function of the PDZ domain of Dishevelled in noncanonical Wnt signalling. Journal of Cell Science, 135(11). View this article in WRRO


- QuantifyPolarity, a new tool-kit for measuring planar polarized protein distributions and cell properties in developing tissues. Development, 148(18). View this article in WRRO


- How do the Fat–Dachsous and core planar polarity pathways act together and independently to coordinate polarized cell behaviours?. Open Biology, 11(2). View this article in WRRO


- Molecular mechanisms mediating asymmetric subcellular localisation of the core planar polarity pathway proteins. Biochemical Society Transactions, 48(4), 1297-1308. View this article in WRRO


- DAnkrd49 and Bdbt act via casein kinase Iε to regulate planar polarity in Drosophila. PLoS Genetics, 16(8).


- Planar Cell Polarity Effector Proteins Inturned and Fuzzy Form a Rab23 GEF Complex. Current Biology, 29(19), 3323-3330.e8.


- Robust Wnt signaling is maintained by a Wg protein gradient and Fz2 receptor activity in the developing Drosophila wing. Development, 146(15).


- Experimental and theoretical evidence for bidirectional signaling via core planar polarity protein complexes in Drosophila. iScience, 17, 49-66. View this article in WRRO


- Reciprocal action of casein kinase Iε on core planar polarity proteins regulates clustering and asymmetric localisation. eLife, 2019(8).


- Retromer controls planar polarity protein levels and asymmetric localization at intercellular junctions. Current Biology, 29(3), 484-491.e6. View this article in WRRO


- A theoretical framework for planar polarity establishment through interpretation of graded cues by molecular bridges. Development, 146(3). View this article in WRRO


- Rapid disruption of dishevelled activity uncovers an intercellular role in maintenance of prickle in core planar polarity protein complexes. Cell Reports, 25(6), 1415-1424.e6. View this article in WRRO


- Integrating planar polarity and tissue mechanics in computational models of epithelial morphogenesis. Current Opinion in Systems Biology, 5, 41-49. View this article in WRRO


- A Dual Function for Prickle in Regulating Frizzled Stability during Feedback-Dependent Amplification of Planar Polarity. Current Biology, 27(18), 2784-2797.e3. View this article in WRRO


- Robust Asymmetric Localization of Planar Polarity Proteins Is Associated with Organization into Signalosome-like Domains of Variable Stoichiometry.. Cell Rep, 17(10), 2660-2671.


- Adhesion GPCRs Govern Polarity of Epithelia and Cell Migration.. Handb Exp Pharmacol, 234, 249-274.


- Planar cell polarity: the Dachsous/Fat system contributes differently to the embryonic and larval stages of Drosophila.. Biol Open, 5(4), 397-408.


- Conservation of Planar Polarity Pathway Function Across the Animal Kingdom.. Annu Rev Genet, 49, 529-551.


- Cellular interpretation of the long-range gradient of Four-jointed activity in the Drosophila wing.. Elife, 4.


- Planar polarity: forcing cells into line.. Curr Biol, 25(21), R1032-R1034.


- Rabaptin-5 and Rabex-5 are neoplastic tumour suppressor genes that interact to modulate Rab5 dynamics in Drosophila melanogaster.. Dev Biol, 385(1), 107-121.


- Control of tissue morphology by Fasciclin III-mediated intercellular adhesion.. Development, 140(18), 3858-3868.


- Strabismus promotes recruitment and degradation of farnesylated prickle in Drosophila melanogaster planar polarity specification.. PLoS Genet, 9(7), e1003654.


- Localised JAK/STAT pathway activation is required for Drosophila wing hinge development.. PLoS One, 8(5), e65076.


- An intracellular partitioning-based framework for tissue cell polarity in plants and animals.. Development, 140(10), 2061-2074.


- A Cul-3-BTB ubiquitylation pathway regulates junctional levels and asymmetry of core planar polarity proteins.. Development, 140(8), 1693-1702.


- The Frizzled-dependent planar polarity pathway locally promotes E-cadherin turnover via recruitment of RhoGEF2. Journal of Cell Science, 126(5), e1-e1.


- The Frizzled-dependent planar polarity pathway locally promotes E-cadherin turnover via recruitment of RhoGEF2.. Development, 140(5), 1045-1054.


- Structure-function dissection of the frizzled receptor in Drosophila melanogaster suggests different mechanisms of action in planar polarity and canonical Wnt signaling.. Genetics, 192(4), 1295-1313.


- Planar polarity specification through asymmetric subcellular localization of Fat and Dachsous.. Curr Biol, 22(10), 907-914.


- The roles of the cadherins Fat and Dachsous in planar polarity specification in Drosophila.. Dev Dyn, 241(1), 27-39.


- Principles of planar polarity in animal development.. Development, 138(10), 1877-1892.


- Dynamics of core planar polarity protein turnover and stable assembly into discrete membrane subdomains.. Dev Cell, 20(4), 511-525.


- Frizzled signaling: Gαo and Rab5 at the crossroads of the canonical and PCP pathways?. Sci Signal, 3(148), pe43.


- Four-jointed modulates growth and planar polarity by reducing the affinity of dachsous for fat.. Curr Biol, 20(9), 803-810.


- Gradients and the specification of planar polarity in the insect cuticle.. Cold Spring Harb Perspect Biol, 1(5), a000489.


- Asymmetric localisation of planar polarity proteins: Mechanisms and consequences.. Semin Cell Dev Biol, 20(8), 957-963.


- The planar polarity pathway.. Curr Biol, 18(19), R898-R902.


- Differential stability of flamingo protein complexes underlies the establishment of planar polarity.. Curr Biol, 18(20), 1555-1564.


- Planar polarity genes in the Drosophila wing regulate the localisation of the FH3-domain protein Multiple Wing Hairs to control the site of hair production.. Development, 135(18), 3103-3111.


- Microcephalin coordinates mitosis in the syncytial Drosophila embryo.. J Cell Sci, 120(Pt 20), 3578-3588.


- The planar polarity pathway promotes coordinated cell migration during Drosophila oogenesis.. Development, 134(17), 3055-3064.


- Differential activities of the core planar polarity proteins during Drosophila wing patterning.. Dev Biol, 302(1), 181-194.


- Planar polarity is positively regulated by casein kinase Iepsilon in Drosophila.. Curr Biol, 16(13), 1329-1336.


- Polarized transport of Frizzled along the planar microtubule arrays in Drosophila wing epithelium.. Dev Cell, 10(2), 209-222.


- Long-range coordination of planar polarity patterning in Drosophila. Advances in Developmental Biology, 14, 39-57.


- Long-range coordination of planar polarity in Drosophila.. Bioessays, 27(12), 1218-1227.


- Organ shape: controlling oriented cell division.. Curr Biol, 15(18), R758-R759.


- Mathematical modeling of planar polarity.. Dev Cell, 8(2), 134-136.


- Cleavage and secretion is not required for Four-jointed function in Drosophila patterning.. Development, 131(4), 881-890.


- EGF signaling and ommatidial rotation in the Drosophila eye.. Curr Biol, 13(16), 1451-1457.


- Frizzled signalling and cell polarisation in Drosophila and vertebrates.. Development, 130(19), 4501-4513.


- Strabismus is asymmetrically localised and binds to Prickle and Dishevelled during Drosophila planar polarity patterning.. Development, 130(13), 3007-3014.


- Nonautonomous planar polarity patterning in Drosophila: dishevelled-independent functions of frizzled.. Dev Cell, 3(6), 851-863.


- Planar polarity: photoreceptors on a high fat diet.. Curr Biol, 12(11), R384-R385.


- Asymmetric localization of frizzled and the determination of notch-dependent cell fate in the Drosophila eye.. Curr Biol, 12(10), 813-824.


- The asymmetric subcellular localisation of components of the planar polarity pathway.. Semin Cell Dev Biol, 13(3), 225-231.


- Planar polarity: getting ready to ROCK.. Curr Biol, 11(13), R506-R509.


- Asymmetric localization of frizzled and the establishment of cell polarity in the Drosophila wing.. Mol Cell, 7(2), 367-375.


- Multiple roles for four-jointed in planar polarity and limb patterning.. Dev Biol, 228(2), 181-196.


- Nuclear signaling by Rac and Rho GTPases is required in the establishment of epithelial planar polarity in the Drosophila eye.. Curr Biol, 10(16), 979-988.


- The four-jointed gene is required in the Drosophila eye for ommatidial polarity specification.. Curr Biol, 9(23), 1363-1372.


- Polarity determination in the Drosophila eye.. Curr Opin Genet Dev, 9(4), 442-446.


- Polarity determination in the Drosophila eye: a novel role for unpaired and JAK/STAT signaling.. Genes Dev, 13(10), 1342-1353.


- Dishevelled activates JNK and discriminates between JNK pathways in planar polarity and wingless signaling.. Cell, 94(1), 109-118.


- Hedgehog is an indirect regulator of morphogenetic furrow progression in the Drosophila eye disc.. Development, 124(17), 3233-3240.


- The role of RhoA in tissue polarity and Frizzled signalling.. Nature, 387(6630), 292-295.


- The regulation of hedgehog and decapentaplegic during Drosophila eye imaginal disc development.. Mech Dev, 58(1-2), 39-50.


- Ommatidial polarity in the Drosophila eye is determined by the direction of furrow progression and local interactions.. Development, 121(12), 4247-4256.


- Regulation of furrow progression in the Drosophila eye by cAMP-dependent protein kinase A.. Nature, 373(6516), 705-709.


- Characterisation of T48, a target of homeotic gene regulation in Drosophila embryogenesis.. Mech Dev, 46(1), 27-39.


- Targets of homeotic gene regulation in Drosophila.. J Cell Sci Suppl, 16, 53-60.


- Targets of homeotic gene control in Drosophila.. Nature, 348(6299), 308-312.


- Characterisation of cell-scale signalling by the core planar polarity pathway during Drosophila wing development. eLife, 14.


Book chapters
- Use of Fluorescence Recovery After Photobleaching (FRAP) to Measure In Vivo Dynamics of Cell Junction–Associated Polarity Proteins, Methods in Molecular Biology (pp. 1-30). Springer US


- Long-range coordination of planar polarity patterning in Drosophila, PLANAR CELL POLARIZATION DURING DEVELOPMENT (pp. 39-57).


Conference proceedings
- Examination of cell signalling involved in the convergent extension movements during Drosophila embryonic hindgut development. MECHANISMS OF DEVELOPMENT, Vol. 126 (pp S176-S176)


- The planar polarity pathway and trachea formation in the Drosophila embryo. MECHANISMS OF DEVELOPMENT, Vol. 126 (pp S80-S80)


- Characterisation of the role of ubiquitin-like molecules in regulating sub-cellular trafficking of core planar polarity proteins in the Drosophila wing. MECHANISMS OF DEVELOPMENT, Vol. 126 (pp S144-S144)


- The University of Sheffield RNAi Screening facility: Genome-wide RNAi screens in Drosophila cells using a 2nd generation in silico optimized library. MECHANISMS OF DEVELOPMENT, Vol. 126 (pp S290-S290)


- Non-canonical Wnt/frizzled signalling and cell polarity in Drosophila. EUROPEAN JOURNAL OF CELL BIOLOGY, Vol. 82 (pp 84-85)


Preprints
- Characterisation of cell-scale signalling by the core planar polarity pathway during Drosophila wing development, eLife Sciences Publications, Ltd.


- Characterisation of cell-scale signalling by the core planar polarity pathway during Drosophila wing development, Cold Spring Harbor Laboratory.


- Recombinant biosensors for multiplex and super-resolution imaging of phosphoinositides, Cold Spring Harbor Laboratory.


- DAnkrd49 and Bdbt act via Casein Kinase Iε to regulate planar polarity inDrosophila, Cold Spring Harbor Laboratory.


- Information Flow in Planar Polarity, Cold Spring Harbor Laboratory.


- Molecular symmetry breaking in the Frizzled-dependent planar polarity pathway. Current Biology, 33(24), 5340-5354.e6. View this article in WRRO
- Grants
-
- Wellcome Trust Senior Fellowship in Basic Biomedical Science: ‘Planar polarity as a model for understanding self-organisation from molecular to tissue scales’
- UKRI/Wellcome Trust – Building Collaboration at the Physics of Life Interface: Dr Alex Fletcher and Prof David Strutt (joint-PIs), Prof Ashley Cadby (co-I), ‘Understanding self-organised tissue patterning across scales’
- Teaching activities
-
Undergraduate:
• BIS218 Advanced Developmental Biology Practical Sessions
- Professional activities and memberships
-
- Member of DBT/Wellcome Trust India Alliance Intermediate and Senior Fellowships Selection Committee (2021–)
- Member of Wellcome Trust Cell and Developmental Biology Expert Review Group (2017–2020)
- Member of Human Frontiers Science Program Grants Committee (2013–2017)
- Visiting scientist Kavli Institute for Theoretical Physics (KITP), University of California, Santa Barbara (2013, 2016, 2023)
- Editorial boards of Current Biology and Development Member of the British Society for Developmental Biology and the Genetics Society