Dr Mark Collins
School of Biosciences
Senior Lecturer in Biological Mass Spectrometry
Deputy Director of the Mass Spectrometry Centre


- Profile
-
Deputy Director of the Mass Spectrometry Centre
Expertise: Proteomics- 2019-present: Senior Lecturer, University of Sheffield
- 2013-2018: Lecturer, University of Sheffield
- 2011-2013: Senior Staff Scientist, Wellcome Trust Sanger Institute
- 2005-2011: Research Associate/Staff Scientist, Wellcome Trust Sanger Institute
- 2001-2005: PhD, Division of Neuroscience, University of Edinburgh
- 2000-2001: Research Assistant, Centre for Liver Disease, Mater Misericordiae University Hospital, Dublin.
- 1996-2000: BSc, Department of Biochemistry, University College Dublin
- Research interests
-
We are interested in how proteins are regulated by post-translational modifications and how cell signalling pathways are perturbed in disease. PTMs such as phosphorylation, ubiquitination, acetylation and palmitoylation regulate overlapping and distinct aspects of protein function, but the interplay of these modifications is not well understood.
We exploit biochemical and quantitative mass spectrometry-based approaches to understand how proteins are dynamically regulated by protein synthesis and degradation and an array of post-translational modifications.
Research themes
Neuroproteomics
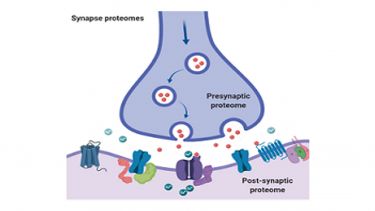
We have a long-standing interest in understanding how brain proteomes relate to fundamental aspects of learning and memory and how they are altered in disease. We have been at the forefront of proteomic studies of synapses from the interrogation of the composition of pre and post-synaptic fractions to the characterisation of postsynaptic glutamate receptor associated protein complexes and their regulation by post-translational modifications.
We are currently investigating how the synapse proteome is regulated by palmitoylation and lysine acetylation; important PTMs that regulate association of proteins with pre and post synaptic membranes and protein stability, respectively.
We are together with our collaborators investigating the composition of synapse proteomes in several mouse models of disease and human iPS cell derived neuronal models of disease including Schizophrenia, Alzheimer's disease and ALS. Furthermore, we are currently conducting clinical proteomics studies of ALS patient cohorts to identify biomarkers of oxidative stress to enable the action of therapeutics to be assessed.
Regulation of membrane protein function by S-acylation (Palmitoylation)
Palmitoylation, the only known reversible lipid modification of proteins, is a critical regulator of protein trafficking, stability and signalling and is important for all cell types, and organisms from yeast to humans.
Proteins can be palmitoylated by a family of 23 protein acyl transferases (PATs) in humans and many of these enzymes have been shown to regulate important aspects of cell biology and in particular in neuronal cells in the brain where palmitoylation of receptors and associated proteins are essential for communication between brain cells and therefore functions such as learning and memory.
Indeed, several PATs have been implicated in the pathophysiology of neurological disorders from Huntington’s disease to intellectual disability and schizophrenia and well as other diseases such as diabetes and cancer.
The identification of determinants of substrate specificity of palmitoyl acyltransferases is an important goal toward understanding how palmitoylation regulates cell function. Development of strategies to enrich, identify and quantify proteins and PTMs is also a major focus of research in our lab.
Perturbed signalling pathways in neurodegenerative diseases
Motor Neurone Disease or Amyotrophic lateral sclerosis (ALS) is a disorder that results in fatal paralysis within a few years of symptom onset. Defects in a growing list of genes are associated with the development of ALS/MND.
Many of these gene defects result in the accumulation of aggregates in cells of patients with ALS and these aggregates are believed to cause neurons to die, resulting in the symptoms of the disease.
Recently, a number of large exome sequencing studies of ALS patients have independently identified several loss of function mutations in protein kinases but it is not known how dysregulation of these kinases might contribute to development of disease.
We are using protein biochemistry, immunofluorescence microscopy and phosphoproteomic approaches to understand how human mutations lead to molecular and cellular changes that contribute to to the pathogenesis of ALS.
Mass spectrometry-based proteomics
The key enabling technology for our research is shotgun proteomics; in which protein samples (whole cell lysates, organelles, protein complexes etc.) are digested with into peptides, separated using nanoflow chromatography and analysed using high-resolution tandem mass spectrometry (Faculty of Science Mass Spectrometry Facility). This approach permits unbiased, quantitative analysis of protein levels and PTMs, with unrivalled accuracy and sensitivity.
We exploit affinity purification and proximity labelling strategies to purify and characterise multiprotein complexes formed by protein kinases and palmitoyl-acyltransferases to identify their substrates as well as regulatory proteins that determine substrate specificity or target these enzymes to different subcellular compartments or membrane microdomains.
Collaborations
Prof Shaun Cowley (University of Leicester)
Dr Daniel Humphreys (University of Sheffield)
Prof Dave Kelly (University of Sheffield)
Dr Stéphane Mesnage (University of Sheffield)
Dr Andrew Peden (University of Sheffield)
Dr Richard Mead (University of Sheffield)
Prof Pamela Shaw (University of Sheffield)
- Publications
-
Show: Featured publications All publications
Featured publications
Journal articles
- Typhoid toxin hijacks Wnt5a to establish host senescence and Salmonella infection. Cell Reports, 42(10). View this article in WRRO


- A potential histone-chaperone activity for the MIER1 histone deacetylase complex. Nucleic Acids Research.


- Transcriptional programs regulating neuronal differentiation are disrupted in DLG2 knockout human embryonic stem cells and enriched for schizophrenia and related disorders risk variants. Nature Communications, 13(1), 27-27.


- Proximity-dependent biotin identification (BioID) reveals a dynamic LSD1-CoREST interactome during embryonic stem cell differentiation. Molecular Omics, 18(1), 31-44.


- Proteomic approaches to study cysteine oxidation: applications in neurodegenerative diseases. Frontiers in Molecular Neuroscience, 14.


- Regulation and function of the palmitoyl‐acyltransferase ZDHHC5. The FEBS Journal, 288(23), 6623-6634.


- Cell‐type specific visualization and biochemical isolation of endogenous synaptic proteins in mice. European Journal of Neuroscience, 51(3), 793-805.


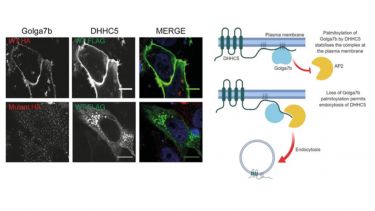
- S‐acylated Golga7b stabilises DHHC5 at the plasma membrane to regulate cell adhesion. EMBO reports, 20(10), ---.


- S-acylation regulates the trafficking and stability of the unconventional Q-SNARE STX19. Journal of Cell Science, 131(20). View this article in WRRO


- Inhibition of somatosensory mechanotransduction by annexin A6. Science Signaling, 11(535). View this article in WRRO


- Arc Requires PSD95 for Assembly into Postsynaptic Complexes Involved with Neural Dysfunction and Intelligence. Cell Reports, 21(3), 679-691. View this article in WRRO


- Global, site-specific analysis of neuronal protein S-acylation. Scientific Reports, 7(1).


- Evolution of complexity in the zebrafish synapse proteome. Nature Communications, 8(-), ---.


- TBK1: a new player in ALS linking autophagy and neuroinflammation.. Mol Brain, 10(1), 5. View this article in WRRO


- A polygenic burden of rare disruptive mutations in schizophrenia. Nature, 506(7487), 185-190.


- De novo CNV analysis implicates specific abnormalities of postsynaptic signalling complexes in the pathogenesis of schizophrenia.. Mol Psychiatry, 17(2), 142-153.


- Characterization of the proteome, diseases and evolution of the human postsynaptic density.. Nat Neurosci, 14(1), 19-21.


All publications
Journal articles
- Protocol to identify mechanosensitive nuclear proteins using tunable actomyosin contractility and proximity biotinylation in mammalian cells. STAR Protocols, 7(1), 104288-104288.


- Dynamics of the CD9 interactome during bacterial infection of epithelial cells by proximity labelling proteomics. The FEBS Journal. View this article in WRRO


- The potential role of misfolded wild-type SOD1 protein in sporadic amyotrophic lateral sclerosis (ALS): a review of the evidence. Neurobiology of Disease, 216, 107124-107124.


- Mutations on the surface of HDAC1 reveal molecular determinants of specific complex assembly and their requirement for gene regulation. Nucleic Acids Research, 53(17). View this article in WRRO


- Glutathione oxidation in cerebrospinal fluid as a biomarker of oxidative stress in amyotrophic lateral sclerosis. Translational Neurodegeneration, 14(1). View this article in WRRO


- Reduced palmitoylation of SQSTM1/p62 in Huntington disease Is associated with impaired autophagy. The FASEB Journal, 39(9). View this article in WRRO


- Mechanical control of the alternative splicing factor PTBP1 regulates extracellular matrix stiffness induced proliferation and cell spreading. iScience, 28(4). View this article in WRRO


- Rapid degradation of histone deacetylase 1 (HDAC1) reveals essential roles in both gene repression and active transcription. Nucleic Acids Research, 53(4). View this article in WRRO


- Regulation of the brain acetylome by Sirtuin-2 and during ageing. Clinica Chimica Acta, 558, 118675-118675.


- #32. Identification of proteins regulating the metastatic spread of breast cancer to bone: Proteomic analysis of the role of the pro-metastatic transcription factor TWIST1 and the gene target micro-RNA (miR-10b). Journal of Bone Oncology, 45, 100570-100570.


- #28. Identification of key proteins within prostate cancer derived exosomes and their role in priming the pre-metastatic niche. Journal of Bone Oncology, 45, 100566-100566.


- FAM81A is a postsynaptic protein that regulates the condensation of postsynaptic proteins via liquid–liquid phase separation. PLOS Biology, 22(3). View this article in WRRO


- The extracellular matrix supports breast cancer cell growth under amino acid starvation by promoting tyrosine catabolism. PLOS Biology, 22(1). View this article in WRRO


- Typhoid toxin hijacks Wnt5a to establish host senescence and Salmonella infection. Cell Reports, 42(10). View this article in WRRO


- A potential histone-chaperone activity for the MIER1 histone deacetylase complex. Nucleic Acids Research.


- Sec22b is a critical and nonredundant regulator of plasma cell maintenance. Proceedings of the National Academy of Sciences, 120(2). View this article in WRRO


- Low expression of EXOSC2 protects against clinical COVID-19 and impedes SARS-CoV-2 replication. Life Science Alliance, 6(1). View this article in WRRO


- The EGFR/ErbB inhibitor neratinib modifies the neutrophil phosphoproteome and promotes apoptosis and clearance by airway macrophages. Frontiers in Immunology, 13.


- Altered subgenomic RNA abundance provides unique insight into SARS-CoV-2 B.1.1.7/Alpha variant infections. Communications Biology, 5.


- Developmental disruption to the cortical transcriptome and synaptosome in a model of SETD1A loss-of-function.. Human Molecular Genetics, 31(18), 3095-3106.


- Transcriptional programs regulating neuronal differentiation are disrupted in DLG2 knockout human embryonic stem cells and enriched for schizophrenia and related disorders risk variants. Nature Communications, 13(1), 27-27.


- Proximity-dependent biotin identification (BioID) reveals a dynamic LSD1-CoREST interactome during embryonic stem cell differentiation. Molecular Omics, 18(1), 31-44.


- PGFinder, a novel analysis pipeline for the consistent, reproducible, and high-resolution structural analysis of bacterial peptidoglycans. eLife, 10.


- Proteomic approaches to study cysteine oxidation: applications in neurodegenerative diseases. Frontiers in Molecular Neuroscience, 14.


- Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition).. Autophagy, 17(1), 1-382. View this article in WRRO


- Regulation and function of the palmitoyl‐acyltransferase ZDHHC5. The FEBS Journal, 288(23), 6623-6634.


- Cell‐type specific visualization and biochemical isolation of endogenous synaptic proteins in mice. European Journal of Neuroscience, 51(3), 793-805.


- Apoptotic signalling targets the post-endocytic sorting machinery of the death receptor Fas/CD95. Nature Communications, 10(1), ---.


- S‐acylated Golga7b stabilises DHHC5 at the plasma membrane to regulate cell adhesion. EMBO reports, 20(10), ---.


- Proteomic profiling, transcription factor modeling, and genomics of evolved tolerant strains elucidate mechanisms of vanillin toxicity in Escherichia coli.. mSystems, 4(4).


- S-acylation regulates the trafficking and stability of the unconventional Q-SNARE STX19. Journal of Cell Science, 131(20). View this article in WRRO


- Inhibition of somatosensory mechanotransduction by annexin A6. Science Signaling, 11(535). View this article in WRRO


- Site specific modification of adeno-associated virus enables both fluorescent imaging of viral particles and characterization of the capsid interactome. Scientific Reports, 7. View this article in WRRO


- Arc Requires PSD95 for Assembly into Postsynaptic Complexes Involved with Neural Dysfunction and Intelligence. Cell Reports, 21(3), 679-691. View this article in WRRO


- Global, site-specific analysis of neuronal protein S-acylation. Scientific Reports, 7(1).


- Evolution of complexity in the zebrafish synapse proteome. Nature Communications, 8(-), ---.


- TBK1: a new player in ALS linking autophagy and neuroinflammation.. Mol Brain, 10(1), 5. View this article in WRRO


- Human post-mortem synapse proteome integrity screening for proteomic studies of postsynaptic complexes. Molecular Brain, 7.


- AMPA Receptor Complex Dynamics in Time and Space. Neuron, 84(1), 1-3.


- Confident and sensitive phosphoproteomics using combinations of collision induced dissociation and electron transfer dissociation.. Journal of Proteomics, 103, 1-14.


- Phosphoinositide metabolism links cGMP-dependent protein kinase G to essential Ca²⁺ signals at key decision points in the life cycle of malaria parasites.. PLoS Biology, 12(3), ---.


- A polygenic burden of rare disruptive mutations in schizophrenia. Nature, 506(7487), 185-190.


- Comparative study of human and mouse postsynaptic proteomes finds high compositional conservation and abundance differences for key synaptic proteins.. PLoS One, 7(10), e46683.


- Analysis of protein palmitoylation reveals a pervasive role in Plasmodium development and pathogenesis.. Cell Host Microbe, 12(2), 246-258.


- A Plasmodium calcium-dependent protein kinase controls zygote development and transmission by translationally activating repressed mRNAs.. Cell Host Microbe, 12(1), 9-19.


- SynGAP isoforms exert opposing effects on synaptic strength.. Nat Commun, 3, 900.


- Enhanced peptide identification by electron transfer dissociation using an improved Mascot Percolator.. Mol Cell Proteomics, 11(8), 478-491.


- De novo CNV analysis implicates specific abnormalities of postsynaptic signalling complexes in the pathogenesis of schizophrenia.. Mol Psychiatry, 17(2), 142-153.


- Coordinating cell cycle progression via cyclin specificity.. Cell Cycle, 10(24), 4195-4196.


- Quantitative proteomics reveals the basis for the biochemical specificity of the cell-cycle machinery.. Mol Cell, 43(3), 406-417.


- Shotgun proteomics aids discovery of novel protein-coding genes, alternative splicing, and "resurrected" pseudogenes in the mouse genome.. Genome Res, 21(5), 756-767.


- Characterization of the proteome, diseases and evolution of the human postsynaptic density.. Nat Neurosci, 14(1), 19-21.


- A comprehensive survey of protein palmitoylation in late blood-stage Plasmodium falciparum. Malaria Journal, 9(S2).


- A comprehensive survey of protein palmitoylation in late blood-stage Plasmodium falciparum. Malaria Journal, 9(S2).


- Cell biology. Evolving cell signals.. Science, 325(5948), 1635-1636.


- Targeted tandem affinity purification of PSD-95 recovers core postsynaptic complexes and schizophrenia susceptibility proteins.. Mol Syst Biol, 5, 269.


- Neurotransmitters drive combinatorial multistate postsynaptic density networks.. Sci Signal, 2(68), ra19.


- Mapping multiprotein complexes by affinity purification and mass spectrometry.. Curr Opin Biotechnol, 19(4), 324-330.


- Evolutionary expansion and anatomical specialization of synapse proteome complexity.. Nat Neurosci, 11(7), 799-806.


- Phosphoproteomic analysis of the mouse brain cytosol reveals a predominance of protein phosphorylation in regions of intrinsic sequence disorder.. Mol Cell Proteomics, 7(7), 1331-1348.


- Analysis of protein phosphorylation on a proteome-scale.. Proteomics, 7(16), 2751-2768.


- Supramolecular signalling complexes in the nervous system.. Subcell Biochem, 43, 185-207.


- Molecular characterization and comparison of the components and multiprotein complexes in the postsynaptic proteome.. J Neurochem, 97 Suppl 1, 16-23.


- Robust enrichment of phosphorylated species in complex mixtures by sequential protein and peptide metal-affinity chromatography and analysis by tandem mass spectrometry.. Sci STKE, 2005(298), pl6.


- Proteomic analysis of in vivo phosphorylated synaptic proteins. Journal of Biological Chemistry, 280(7), 5972-5982.


- Polymorphisms in tumour necrosis factor-alpha, transforming growth factor-beta, interleukin-10, interleukin-6, interferon-gamma, and outcome of hepatitis C virus infection.. J Med Virol, 71(2), 212-218.


- Typhoid toxin of Salmonella Typhi elicits host antimicrobial response during acute typhoid fever. EMBO Molecular Medicine.


- APC15 drives the turnover of MCC-CDC20 to make the spindle assembly checkpoint responsive to kinetochore attachment.. Nat Cell Biol, 13(10), 1234-1243.


- Analysis protein complexes by 1D-SDS-PAGE and tandem mass spectrometry. Protocol Exchange.


Book chapters
- Quantitative analysis of protein S-acylation site dynamics using site-specific acyl-biotin exchange (ssABE). In Evans C, Wright P & Noirel J (Ed.), Mass Spectrometry of Proteins (pp. 71-82). New York: Humana Press. View this article in WRRO


- Neuroproteomics., Encyclopedia of Neuroscience (pp. 971-981). Oxford: Academic Press


- Neuroproteomics, Encyclopedia of Neuroscience Volumes 1 11 (pp. V7-V7-981).


- Supramolecular signalling complexes in the nervous system, Subcellular Proteomics


- Proteomics in the Neurosciences, Proteomics: Biomedical and Pharmaceutical Applications Kluwer Academic Press


Conference proceedings
- Site specific labelling of adeno-associated virus identifies targets for enhancing viral transduction efficiency. Human Gene Therapy, Vol. 27(7) (pp A14-A14). London, UK, 15 June 2016 - 15 June 2016. View this article in WRRO


- Gene Therapy for Familial ALS Using AAV9 Mediated Silencing of Mutant SOD1. Human Gene Therapy, Vol. 27(7) (pp A12-A12). University College London Institute of Child Health View this article in WRRO


- Resolving Sites of Protein Palmitoylation Using Quantitative Proteomics. MOLECULAR & CELLULAR PROTEOMICS, Vol. 13(8) (pp S41-S41)


- APC15 drives the turnover of MCC-Cdc20 to make the spindle assembly checkpoint responsive to kinetochore attachment. FEBS JOURNAL, Vol. 279 (pp 556-556)


Preprints
- Secondary nucleation of α-Synuclein drives Mitochondria dysfunctions and Lewy body formation in Parkinson’s Disease, Springer Science and Business Media LLC.


- An unbiased screening approach for identifying proteins with a mechanosensitive nuclear localization, Cold Spring Harbor Laboratory.


- Secondary nucleation of α-Synuclein drives Mitochondria dysfunctions and Lewy body formation in Parkinson’s Disease, Cold Spring Harbor Laboratory.


- Mutations on the surface of HDAC1 reveal molecular determinants of specific complex assembly and their requirement for gene regulation, Cold Spring Harbor Laboratory.


- Typhoid toxin of Salmonella Typhi elicits host antimicrobial response during acute typhoid fever, Cold Spring Harbor Laboratory.


- Dynamics of the CD9 interactome during bacterial infection of epithelial cells by proximity labelling proteomics, Cold Spring Harbor Laboratory.


- Rapid degradation of Histone Deacetylase 1 (HDAC1) reveals essential roles in both gene repression and active transcription, Cold Spring Harbor Laboratory.


- Mechanical control of the splicing factor PTBP1 regulates extracellular matrix stiffness-induced cell proliferation and mechanomemory, Cold Spring Harbor Laboratory.


- Reduced S-acylation of SQSTM1/p62 in Huntington disease is associated with impaired autophagy, Cold Spring Harbor Laboratory.


- Typhoid toxin hijacks Wnt5a to potentiate TGFβ-mediated senescence and Salmonella infections, Cold Spring Harbor Laboratory.


- An unexpected histone chaperone function for the MIER1 histone deacetylase complex, Cold Spring Harbor Laboratory.


- The EGFR/ErbB inhibitor neratinib modifies the neutrophil phosphoproteome and promotes apoptosis and clearance by airway macrophages, Cold Spring Harbor Laboratory.


- Low expression of EXOSC2 protects against clinical COVID-19 and impedes SARS-CoV-2 replication, Cold Spring Harbor Laboratory.


- Plasma cell maintenance and antibody secretion are under the control of Sec22b-mediated regulation of organelle dynamics, Cold Spring Harbor Laboratory.


- LRRK2-mediated phosphorylation of HDAC6 regulates HDAC6-cytoplasmic dynein interaction and aggresome formation, Cold Spring Harbor Laboratory.


- Cell-type specific visualization and biochemical isolation of endogenous synaptic proteins in mice, Cold Spring Harbor Laboratory.


- Regulation of somatosensory mechanotransduction by Annexin A6, Cold Spring Harbor Laboratory.


- PGfinder, a novel analysis pipeline for the consistent, reproducible and high-resolution structural analysis of bacterial peptidoglycans.


- Typhoid toxin hijacks Wnt5a to establish host senescence and Salmonella infection. Cell Reports, 42(10). View this article in WRRO
- Research group
-
PhD students
- Hatoon Alamri
- Weronika Buczek
- Ella Hudson*
- Sam Lewin*
- Ola Shehata*
- Salma Srour*
- Ashley Griffin*
- Ankur Patel*
- Nick Verber*
- Jorge Miguel Ferreira*
*Co-supervisor
- Grants
-
- BBSRC
- MRC
- Weston Park Hospital Cancer Charity
- Royal Society
- Teaching activities
-
Undergraduate and postgraduate taught modules
Undergraduate:
- BMS109 Cell & Molecular
- Level 3 Practical and Dissertation Modules
Doctoral Development Programme
- BMS6007: Mass Spectrometry-based Proteomics and Metabolomics Course (Director)
- Professional activities and memberships
-
- Guest Editor at Frontiers in Molecular Neuroscience
- Fellow of the Higher Education Academy (2016)
- Peer reviewer for scientific journals and grant-awarding bodies
- Opportunities
I welcome applications from international students who are self-funded or have (or are applying for) a government scholarship.
We advertise PhD opportunities (Funded or Self-Funded) on FindAPhD.com
For further information and details of other projects on offer, please see the department PhD Opportunities page.